miRNAs
Plants
Stages
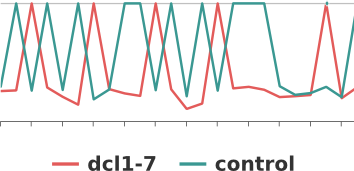
Mutants
NGS samples
Overview
mirEX2 is a comprehensive platform for comparative analysis of primary microRNA expression data. RT–qPCR-based gene expression profiles are stored in a universal and expandable database scheme and wrapped by an intuitive user-friendly interface.
Main features
- User-friendly graphical querying of pri-miRNA/miRNA expression data
- Comparative browsing of the expression levels across plant species, stages, tissues and mutants
- Clear pictograph representation of pri-miRNA expression levels
- Real time data graphical visualization and threshold value selection
- One-click RT-qPCR & NGS data integration
- Dynamic graphs for miRNA biogenesis mutants expression analysis
- Customized download of any browsing result
- Degradome data for selected plant species
Plants growing conditions
Arabidopsis thaliana (L.) Heynh., Columbia-0 wild-type plants, were grown in ‘Jiffy-7 42mm’ soil (Jiffy International AS, Kristiansand, Norway) in a growth chamber with a 16 h day (130–150 µmol m-2 s-1), constant temperature 22°C and humidity of 70%. 10 day-old seedlings were grown in MS medium under the same conditions as above.
Primers design
For each miRNA gene two primers were designed to amplify the fragment of pri-miRNA which contains a mature miRNA. The first step of primer designing was to determine specific regions of each miRNA precursor, using Megablast algorithm. To designe primers we used Primer Express 3.0 and Primer 3.0.4.0. All oligonucleotides fulfil following conditions: 20-27 nucleotide in length, melting temperature between 59 and 61°C, and the amplicon size 120-250 base pairs. We used the PerlPrimer software to avoid self-pairing between primers and the possibility of secondary structure formation.
cDNA preparation
Total RNA was isolated using TRIzol Reagent (Invitrogen by Life Technologies). In the case of siliques and seeds total RNA isolations were carried out as described in [Onate-Sanchez L. and Vicente-Carbajosa J. (2008)]. RNA concentration was measured using a NanoDrop ND-1000 (NanoDrop Technologies). DNA was removed by digestion using TURBO DNA-free kit (Ambion by Life Technologies). Reverse transcription (RT) was performed using 2-3 µg of RNA, Oligo(dT)15 Primer (Novazyme) and SuperScript III Reverse Transcriptase (Invitrogen by Life Technologies) according to the manufacturer’s instructions. After each RT reaction RNA integrity and processivity of reverse transcriptase were checked as described previously by Czechowski [Czechowski T. et al. (2005)].
Real-time PCR analysis
Real-time PCRs were performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems by Life Technologies) and PowerSYBR Green to monitor dsDNA synthesis. The reaction mixture (10µ) contained 5 µl of 2x PowerSYBR Green PCR Master Mix (Applied Biosystems by Life Technologies ), cDNA and gene-specific primers (200 nM each). The following thermal profile was used for all real-time PCRs: 95ºC for 10 min; 40 cycles of: 95ºC for 15 s, and 60ºC for 1 min. After each real-time PCR run, dissociation curve analyses were performed. The results were analyzed using the SDS 2.2.1 software (Applied Biosystems by Life Technologies). Ct values for all miRNA transcripts were normalized to the ACT2 (at3g18780) and PP2AA3 (at1g13320) Ct value. The R2 values were calculated using LinReg v 11.1 software [Ruijter J.M. et al. (2009), Ramakers,C. et al. (2003)].
Northern blot hybridizations
RNA (25 µg) was fractionated on a 15% denaturing polyacrylamide gel (PAGE), transferred to a Hybond-XL membrane (Amersham Biosciences-GE Healthcare, Little Chalfont, UK) using Trans-Blot® Electrophoretic Transfer Cell (Bio-Rad) , and fixed by UV-crosslinking. Pre-hybridization was carried out at 42ºC for 30 min using PerfectHyb Hybridization Buffer (Sigma, Deisenhofen, Germany). Probes were labeled with [32P] γATP (6000 Ci/mmol; NEN-PerkinElmer Life and Analytical Sciences, Waltham, Massachusetts, USA) using T4 polynucleotide kinase (Roche, Mannheim, Germany) and purified on Illustra MicroSpin G-25 Columns (GE Healthcare). Hybridization was performed overnight at 42ºC. Oligonucleotide DNA probes were complementary to miRNA mature molecules, respectively. A probe complementary to U6 snRNA (50 TCATCCTTGCGCAGGGG CCA 30 ) was used as a loading control.
mirEX 2.0 NGS data processing procedures
The adaptor sequences were identified and trimmed by a customized Perl script. Reads in which the adaptor could not be identified were discarded. Additionally, low complexity reads composed in more than 75% of the same nucleotide were discarded. This threshold was set according to sequence composition of known mature plant miRNAs deposited in miRBase [Griffiths-Jones, 2004]. Bowtie program was used to align the reads from each sRNA-seq library to plant mature miRNAs from miRBase v20 [Griffiths-Jones, 2004, Langmead et al., 2009] with up to two mismatches. The raw counts of reads for each library were normalized by the total number of sequences matching miRNA and presented as “reads per million” (RPM).
If you make use of the data presented here, please cite the following articles in addition to the primary data sources:
- Zielezinski A et al. (2015)
mirEX 2.0 - an integrated environment for expression profiling of plant microRNAs.
BMC Plant Biol. 16;15:144. [PubMed ]
Recent Updates
include miRNA biogenesis mutants and plant species representing agronomically significant and comparatively attractive organisms.
New tools for database content exploration allowing for graphical query building and comparative studies.